Looking at the creation time of this draft, it was actually June 2024… Now it’s June 2025, and I suddenly understand why so many content creators become “pigeons” (procrastinators). Starting projects is fun, but finishing them is painful… This is one of my few pure bioinformatics posts…
The problem originated last year when I needed to run monocle3 for pseudotime analysis, but encountered an annoying issue at the final stage. In monocle3, the starting point for pseudotime trajectory needs to be manually specified by the analyst. During R code execution, it automatically opens a browser where users need to specify the starting point on a temporary webpage, then close the page for the analysis to continue.
However, Jupyter’s irkernel doesn’t support this feature. This means I couldn’t complete the analysis directly in Jupyter notebook. This issue was first reported in 2019, but even by 2024 when I needed to do the analysis - five years later - there was still no solution…
But I did accidentally discover a solution when GitHub recommended me a project called jupyter-xeus. This project aims to develop a new series of Jupyter kernels, including a new R kernel called xeus-r. This new kernel supports interactive mode and can generate a link when running monocle3. Users can click the link to complete point selection, then exit the page to continue analysis.
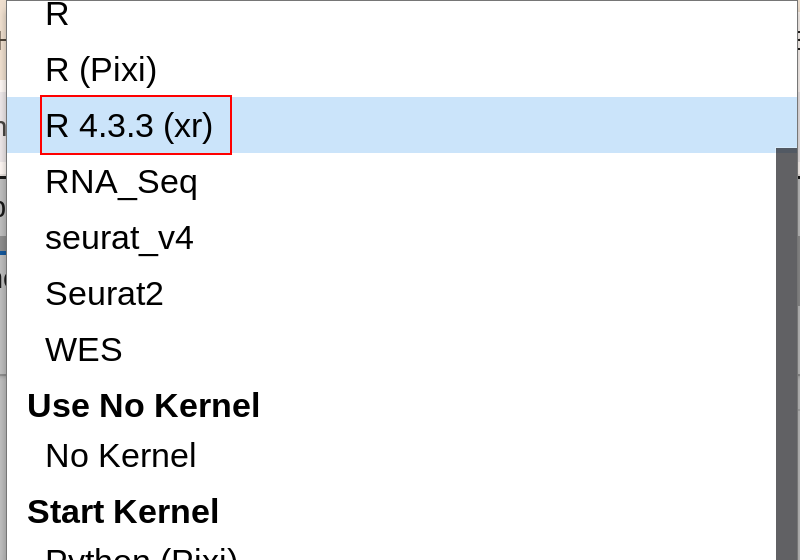
xeus-r is available on conda and can be quickly installed using conda/mamba/pixi. After installation, you’ll see the corresponding xr kernel in Jupyter’s interface - just select it to use.
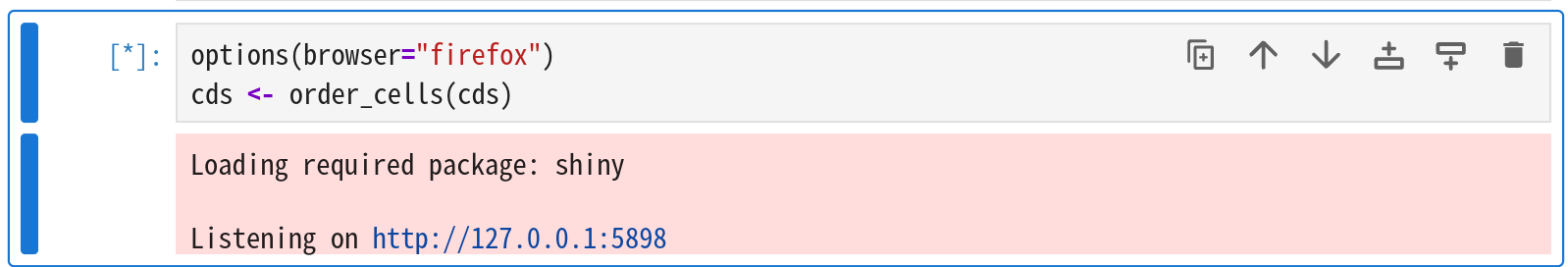
The code to execute is:
1 | library(monocle3) |
After running, you’ll see the link to open below the cell. Open it, select the starting point, then close it to continue.