Lumpy’s author positioned this project more as building a general structural variant signal generation scheme rather than creating an excellent tool. It was implemented as a basic version of the scheme. Compared to other tools, Lumpy spent more time in its article explaining the evidence concepts and methods for obtaining information. Interpreting this tool has made me understand better how to obtain structural variant signals from NGS sequencing data.
Tool Basics
Lumpy is written in C++ and all its functionalities seem to be implemented independently without referencing any existing modules. The software can be obtained via conda, and the program itself hasn’t been updated for several years, so there’s no need to worry about compiling from source code.
The output format of Lumpy results can be selected between VCF and BEDPE. However, it’s worth noting that these two formats contain different information; VCF is more detailed. Additionally, this tool relies entirely on the alignment records in BAM files. Therefore, if the alignment itself is not ideal, it will inevitably affect downstream results. Furthermore, the tool does not contain any random components; with the same input files, the detection results will be identical.
Lumpy’s Structural Variant Model
Lumpy first created a structural variant breakpoint model: $b = (E,l,r,v)$
In this model, breakpoints are defined as pairs of genes that are adjacent in the target genome but not in the reference genome. These pairs of breakpoints are described by four indicators:
- $E$: A set of evidence supporting the breakpoint
- $l$: The left breakpoint interval (interval), containing a vector p representing the relative probability of each point being the true breakpoint position
- $r$: Consistent with $l$, containing all information about the right breakpoint
- $v$: Information about the complexity of the breakpoint, mainly containing direction information from different evidence. This indicator is included to retain more original information because when results are detected, they will only be labeled as one type of structural variant (e.g., deletion), but the original signal may contain support for other types of evidence.
Lumpy uses both SR and RP signals, each parsing out an instance of a breakpoint. After reading all breakpoint instances, it needs to infer whether the same breakpoint exists based on the information contained and then merge breakpoints. The conditions for merging are:
- Both sides of the breakpoints have overlapping regions (threshold unspecified)
- Consistent complexity
If these conditions are met, the two breakpoints can be merged. $E$ will contain the original signals from both breakpoints, $v$ remains unchanged (since it is consistent), and $l$ and $r$ are combined by taking the arithmetic mean. That is, the start of $l$ is the average of the starts of $l1$ and $l2$, and the end is also the average of the ends of $l1$ and $l2$, $r$ is similar (but when there are multiple, how it executes specifically is unknown; taking averages two by two results in different outcomes):
$$merge.start = \frac{l1.start + l2.start}{2}\ \ merge.end = \frac{l1.end + l2.end}{2}$$
According to the author’s description, this method is used to reduce the impact of outlier evidence on the breakpoint interval. After merging the evidence, breakpoints with more than a preset threshold will be detected as structural variants. To determine detected structural variants, information from the breakpoint instances needs to be considered. Here are the default calculation schemes:
- The possible interval of the variant
- The probability that each point in the interval is the true breakpoint
First, the variant’s possible interval (interval) takes the maximum value of the left breakpoints and the minimum value of the right breakpoints (i.e., the smallest possible interval).
While the probability for each point in the interval can be calculated as follows (taking the left breakpoint as an example):
$$l.p[i] = \prod_{e{\in}E}e.l.p[i-o]$$
- $i$ is a certain gene coordinate position
- $l.p$ is the vector of probabilities that a single point is the true breakpoint, with the prefix $l.$ representing the left breakpoint
- $E$ is the set of all evidence
- $e$ is each piece of evidence in the evidence set
- $e.l.p$ is the probability vector for the left breakpoint in a single piece of evidence
- $o$ is the offset value, $o = e.l.s - s.l.s$, i.e., the difference between the start position of the current evidence’s left breakpoint and the predicted start position of the structural variant.
$e.l.p[i-o]$ might seem a bit confusing, but in essence, it’s similar to coverage. The final probability for each point in the interval is the product of the probabilities from all pieces of evidence that have this point. So, theoretically, the closer a point is to having more overlapping regions with original evidence intervals, the smaller its p-value will be, indicating a higher likelihood of being the true breakpoint position.
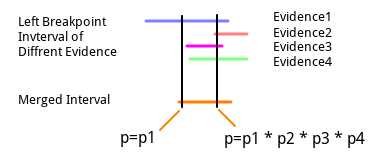
During evidence merging, since the start and end points are continuously averaged to determine the merged interval’s start and end points, it is possible that there will be no interval between the final start and end points (equal or reversed? No instances have been verified). In such cases, the author identifies the maximum point among all sum distributions in $b.E$, removes any distribution not intersecting this point, and processes the resulting subset normally.
Thus, a final result entry is formed. The final result entry’s breakpoint interval can also be reduced according to a preset percentage, for example, taking only the top 99.9%.
Besides the above scheme, there is a Summation scheme used for low-quality data, which will not be interpreted here.
From the above schemes, it is clear that Lumpy essentially reads information-containing alignment records from BAM files, extracts necessary information, and then interprets structural variants based on this information. A crucial point is extracting which information from the signals. To define the required information, Lumpy designed a called Breakpoint evidence interface or commonly used evidence class:
This interface requires three pieces of information:
- is_bp: Whether it contains breakpoint position information
- get_v: Structural variant type information
- get_bpi: Breakpoint possible interval information
That is, for each signal/evidence, the above three pieces of information are interpreted and then added as a separate evidence $e$ to the evidence set $E$ for further interpretation.
So, how does one interpret the above information from signals/evidences? The article explains this with examples from SR, RP, and general breakpoint interface:
RP Evidence Interpretation
In paired-end sequencing, the insert fragment length of Reads Pair has a normal range, and the directions of Read1 and Read2 should be +/-. Any abnormal alignment indicates the presence of a breakpoint. Therefore, the interpreted information is as follows:
- is_bp: Yes, it can determine that a breakpoint exists
- get_v: When two reads are on the same chromosome, based on the actual alignment direction of Read1 and Read2, the mutation type (DEL/INV/DUP) can be inferred. Otherwise, it will be uniformly labeled as
inter-chromosomal - get_bpi: The final breakpoint interval and the p values for each point are obtained through the alignment information of two reads. It is somewhat complex; see below.
Obtaining the breakpoint interval essentially involves mapping the left (l) and right (r) breakpoints from one read’s alignment information to determine the l (containing the interval and the midpoint p value) and r. For example, for l:
Determine whether the breakpoint is upstream or downstream based on the alignment direction of the read.
Before making a judgment, the program estimates the distribution of insert fragment lengths from BAM files without alignment issues to get the average length $l$ and standard deviation $s$ (note that it’s the standard deviation Standard Deviation, not standard deviation). Additionally, a user-specified variable $v$ is required, with $l + vs$ as the expected value for normal insert fragment lengths. Since breaks are unlikely to result in fragments exceeding this expected length, it can be inferred that the breakpoint should be within the range of $l + vs$ (bp) upstream or downstream of the alignment position. This determines the interval range.
For each point $i$ in the above interval, its probability of being a true breakpoint is calculated as $P(S(y).e-S(x).s > i - R(x).s)$ (the original text does not clearly explain what $S()$ represents; I infer it’s a sample abbreviation, with $R()$ representing actual fragment information).
- $y$ represents Read2, and $x$ corresponds to Read1.
- $S(y).e$ is the end position of Read2 in the actual fragment, and $S(x).s$ is the start position of Read1 in the actual fragment.
- $S(y).e-S(x).s$ is the length of the actual fragment.
- $i-R(x).s$ is the distance from point $i$ to the start position of Read1’s alignment.
- $P()$ should be a function that gets the p value from the distribution of normal alignment read fragment lengths. For example, $P(Z>2.94)$ in a standard normal distribution gives the area under the curve, allowing you to evaluate the probability that point $i$ is the true breakpoint.
SR Evidence Interpretation
In SR evidence, one read’s adjacent parts align to non-adjacent genomic positions. In Lumpy, if a read has more than two small fragments, these small fragments are split into pairs of smaller segments for consideration (e.g., three fragments are divided into $x_1,x_2$ and $x_2,x_3$ groups). The information parsed from each pair of small fragments is as follows:
- is_bp: Yes, it can determine that a breakpoint exists
- get_v: When two fragments have the same alignment direction, the structural variant might be DEL or DUP (depending on whether the alignment directions are +/-. If the alignment directions are different but on the same chromosome, it’s INV. If they align to different chromosomes, it’s
inter-chromosomal. - get_bpi: The interval position of the breakpoint is related to the inferred structural variant type. Considering potential sequencing errors, the SR evidence gives a range for the breakpoint position. This range size depends on the input parameter $v$, sample quality (?), and the algorithm used for alignment (?). Since the description in the literature about this part is not as detailed as that of RP, here are some personal speculations; further testing may be needed to confirm:
- About interval acquisition: Ideally, the two parts of a Split-Read断裂 should be non-overlapping. However, in practice, there might be overlapping between the two parts. From Lumpy’s own SR Read extraction script, it indeed allows for some overlap between the two parts. So, the statement that the interval range of SR evidence is related to sample quality and alignment algorithm may refer to this.
- About p value determination within the interval: The author only mentioned that the midpoint p value is the smallest, with p values increasing closer to the edges. It might be a proportional distribution… This would need further testing by creating evidence BAM files.
general breakpoint interface Interpretation
This type of evidence essentially records structural variant information in BEDPE files. Since BEDPE files originally record both sides of the breakpoint interval and the structural variant type, these can be directly mapped to get_v and get_bpi. The description of the BEDPE format will be discussed later; for now, it’s omitted.
The existence of this class of evidence serves two purposes: first, in genetic analysis, it makes it easier to detect variants that are known to exist in a population. Second, when using multiple tools simultaneously, it allows reliable variants detected by other tools to be merged with Lumpy results.


